| Forward Search in Generalized Linear Models | Forward Library Help
|

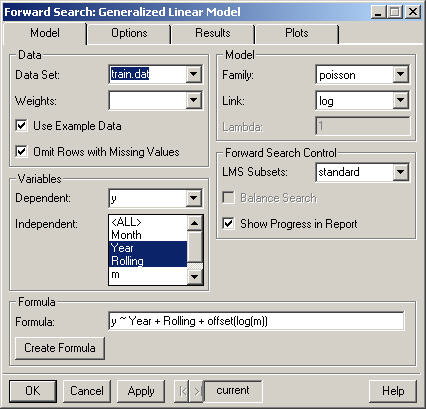
The data to be used in the Forward Search. By default only the data frames in the current working directory are displayed. However, any data frame on the search path (for instance fuel.frame) may be selected by typing its name into the Data Set combo box.
Select the column that specifies weights to be applied to all observations used in the analysis. To weight all rows equally, leave this blank.
Selecting this option causes the Data Set combo box to list the data frames containing the example data provided with the Forward Library. Additionally, when this option is selected the formula, family and link will be filled in automatically.
If this option is selected, rows in the data frame that contain missing values (NA's) will be removed. Otherwise an error will be generated if missing values are present.
Specifies the dependent variable in the model formula.
Specifies the independent (explanatory) variable(s) in the model formula. Use control-clicks to select multiple variables. For more compilcated models select the data (in the Data Set combo box) and press the Create Formula button.
Select the distribution Family for the Model.
Select the link function for the model. The link function of the response is modeled as the sum of linear terms. The possible link functions depend on the family.
The value of the transformation parameter lambda for the Gamma family with Box-Cox link.
The initial subset for the Forward Search in Generalized Linear Models is found by fitting the model with the Forward Library function lmsglm. This option allows the user to control how many subsets are used in the Least Median of Squares criterion. The choices are standard (which uses 100 subsets) and all. Additionally, a specific number of subsets may be specified by typing a number in to the LMS Subsets combo box.
This option is only available for models with binary response. Balancing the search causes the success/failure ratio in each subset of the search to be held as close as possible to the success/failure ratio for the entire data.
Prints a message in the report window for every ten iterations completed in lmsglm (the initial robust estimate) and in the forward search.
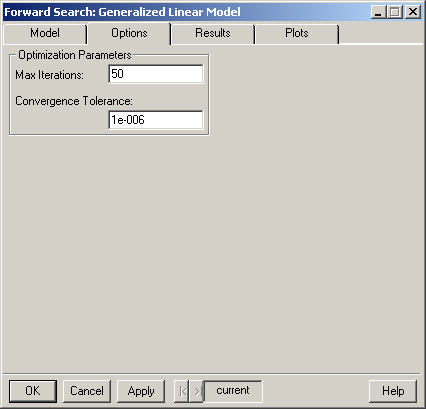
Enter an integer value specifying the maximum number of iterations to perform for the maximum likelihood estimation procedure. If convergence has not been reached after this number of iterations, the procedure stops. The default value appears in the field.
Enter a positive number used as the tolerance for the convergence criterion in the algorithm. This relative offset criterion measures the numerical imprecision in the parameter estimates compared to the statistical variability. Smaller values of Convergence Tolerance require more iterations while larger values result in convergence being declared earlier. The default value appears in the field.
The default values for these parameters are different than those used by the function glm which uses 10 for the maximum number of iterations and 1e-4 for the convergance tolerance.
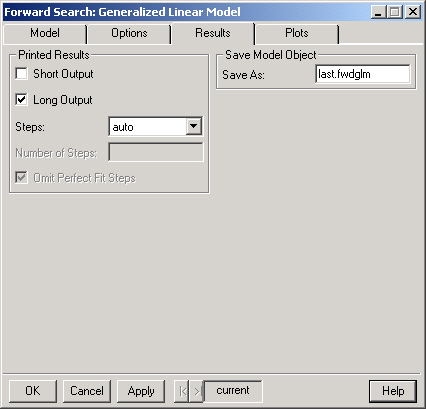
This will display a brief summary of the Forward Search.
This option provides a more detailed summary of the Forward Search. The last Steps (see below) of the diagnostic statistics monitored during the search are displayed.
Controls how many steps are shown in the summary when Long Output is selected. The default behavior ("auto") is to show either the last 5 steps or the last 10% of the search, which ever is greater. Selecting "all" will display the statistics for the entire search and selecting "user" will allow you to enter the desired number of steps in the Number of Steps field.
If the "user" option is selected in Steps then the desired number of Steps should be entered here.
Selecting this option causes the Long Output to omit steps for which there is a perfect fit. Note that a perfect fit can only occur in a model with a binary response.
The Forward Search is saved as an S object with (S version 3) class "fwdglm" in the current working directory. If you do not wish to save the results simply leave this field blank.
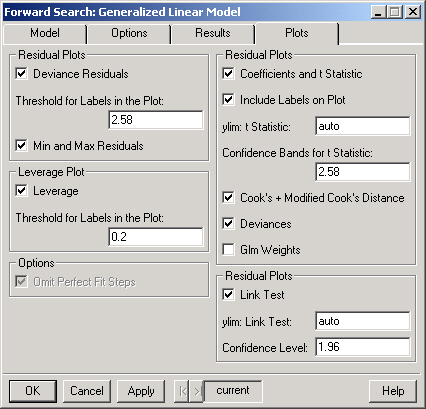
Plots the deviance residuals at each step of the forward search. Note that each line in the plot corresponds to the residual for one observation.
Only observations whose deviance residual exceeds this threshold (at some point during the forward search) will be labeled in the Deviance Residuals plot.
Plots the maximum deviance residual in the subset and the mth overall ordered deviance residual as well as the minimum deviance residual in the complement of the subset and the (m+1)th overall ordered deviance residual for each step in the forward search.
Plots the leverage for each step of the forward search.
Selecting this option causes the plots to omit steps for which there is a perfect fit. Note that a perfect fit can only occur in a model with a binary response.
Plots the coefficients and t statistics for each step of the forward search.
If TRUE then the lines in the coefficients and t statistics plots will be labeled.
The range for the plot of the t statistics. This should be a numeric vector of length 2 containing the lower and upper bounds for the plot. For example, "c(-3.5, 3.5)" (without the quotes) would set the range to show values between -3.5 and 3.5.
The value (on the scale of the t statistics) used to draw the confidence interval on the plot of the t statistics.
Plots the Cook's and modified Cook's for each step of the forward search.
Plots the deviance, the deviance explained, the dispersion parameter and the pseduo R-squared for each step of the forward search.
Plots the weights from the generalized model fit during each step of the forward search.
Plots the goodness of link test at each step of the forward search.
The range for the goodness of link test plot. This should be a numeric vector of length 2 containing the lower and upper bounds for the plot. For example, "c(-10, 10)" (without the quotes) would set the range to show values between -10 and 10.
The value used to draw the confidence interval on the goodness of link test plot.